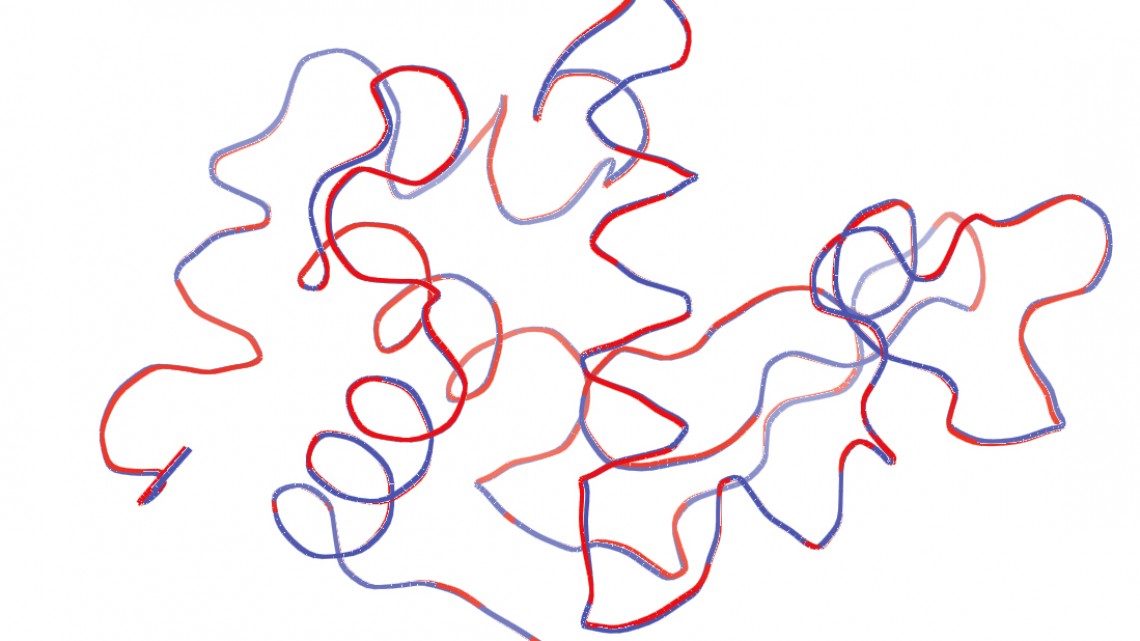
Ribbon representations of protein structure chains derived from both low-intensity "waste" data (blue) and higher-intensity data (red).
Garbage to gold: getting good results from bad data
By Tom Fleischman
A team led by physics professors Sol Gruner and Veit Elser began their recent research by seeking data other researchers had discarded as unusable.
Crazy, you say? To prove their idea was valid, the Cornell scientists needed data that was deemed too unclear – or “noisy” – to be used. The scientists who originally acquired the data were only able to use the best images – about 5 percent of the hundreds of thousands they collected – and threw the rest away. The Cornell group proved that these “garbage” images actually were golden.
Gruner, the John L. Wetherill Professor of Physics and former director of the Cornell High Energy Synchrotron Source (CHESS), aimed to prove that one didn’t need the expense and time of an X-ray free electron laser (XFEL) source to obtain usable protein structure images from many microcrystals. Even with the minimal information gleaned from many incomplete microcrystal diffraction patterns, he said, one can extract the data necessary to paint a complete picture.
The researchers used a method based on the expand-maximize-compress (EMC) algorithm to solve the protein structure of hen egg white lysozyme microcrystals, based on the thrown-away images obtained at Argonne National Laboratory’s Advanced Photon Source(APS).
The group’s work is detailed in “Solving Protein Structure From Sparse Serial Microcrystal Diffraction Data at a Storage Ring Synchrotron Source,” published online July 25 in the International Union of Crystallography Journal. Ti-Yen Lan, Ph.D. ’18, from the Elser Group, is lead author.
Crystals are solids in which atoms form a periodic (ordered) arrangement. They’re everywhere in nature, and crystallography is a scientific technique that uses crystals to determine a material’s molecular structure. A deep understanding of a material’s crystal structure has contributed to major advances in areas including materials science and drug development.
But obtaining suitably large crystals for analysis has been a bottleneck for decades. An alternative is to use data from many tiny crystals. This hasn't been feasible with really small crystals at readily available synchrotron (ring) X-ray sources, but works at XFELs, where an X-ray laser hits the crystal for a matter of a few femtoseconds (quadrillionths of a second), long enough to collect information before the crystal is destroyed by the beam.
XFEL sources are few and far between: There are just four suitable sources in the world, including the Linac Coherent Light Source at Stanford University, the only one in the U.S.
They’re also expensive to operate, Gruner said. “They can only do a few experiments at a time,” he said, “and the operation of the XFEL means the cost per experiment can be the better part of a million dollars.”
Synchrotron ring sources like CHESS or the APS, on the other hand, offer multiple beam lines to run experiments for a small fraction of the cost of XFEL time. The downside: Beams from synchrotron sources hit the material longer – picoseconds (trillionths of a second) instead of femtoseconds – and the radiation damage to the crystal occurs before high-quality X-ray scattering information can be obtained.
At least that’s been the conventional wisdom, Gruner said, noting, “Veit [Elser] and his students have worked out a way to get around that.”
The method involves the EMC algorithm, which models the orientation of each frame probabilistically and reconstructs a consistent 3D intensity model using all the data frames simultaneously. In other words, it takes all the images together and comes up with a 3D reconstruction consistent with all of them.
Even though each individual image on its own is noisy and not interpretable, the images processed with the algorithm together painted a sharp picture of the protein structure. Gold from garbage, indeed.
“What that really means,” Gruner said, “is that the thing that’s been holding up microcrystallography of this type – [not being able to use] synchrotron sources – is no longer a barrier.”
Gruner said lead author Lan has made his methods open-access to benefit scientists the world over.
Other authors included senior research associates Mark Tate and Hugh Philipp of the Cornell Laboratory of Atomic and Solid State Physics; Jennifer Wierman, postdoctoral researcher at MacCHESS (Macromolecular X-ray science at CHESS); and assistant research scientist Jose Martin-Garcia and colleagues at Arizona State University, and Robert Fischetti and colleagues at the APS, who provided the raw data.
This work was supported by grants from the Department of Energy, the Taiwanese government (to Lan), the National Institutes of Health and the National Science Foundation.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe
