'Shield' gives tricky proteins a new identity
By Anne Ju
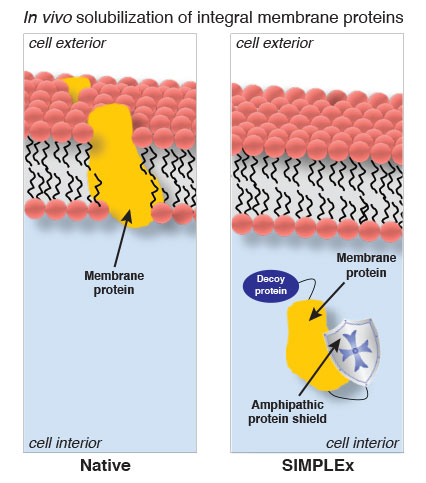
Studding the lipid membrane of every cell are millions of integral membrane proteins – molecules that play crucial roles in cell signaling, adhesion and other life processes, and become the targets for half of today’s pharmaceutical drugs.
Integral membrane proteins also are notoriously hard to synthesize and study, which explains why so few have been fully, three-dimensionally characterized with protein crystallography. Using standard recombinant DNA techniques and some novel design principles, Cornell chemical engineers have developed a new method for making large quantities of integral membrane proteins simply and inexpensively – all without the use of harsh chemicals, or detergents, typically used today.
The method, which they call “SIMPLEx” (Solubilization of Integral Membrane Proteins with high Levels of Expression), could lead to increased characterization and understanding of this all-important class of proteins, and also could be used to make protein machinery for synthetic biological processes. Published online April 8 in Nature Communications, the work was led by Matthew DeLisa, the William L. Lewis Professor of Engineering in the School of Chemical and Biomolecular Engineering, with first author and postdoctoral associate Dario Mizrachi, who designed and carried out the experiments.
Most known protein structures today live inside the cell and are water soluble – happy in the cytoplasmic aqueous environment. But membrane proteins are hydrophobic – they hate water – which is why they’re happiest when embedded in the waterproof lipid bilayer of a cell. Soluble proteins are much easier to make in sufficient quantities for characterization and study.
Making enough membrane proteins to study involves a method called detergent extraction, which employs a chemical to strip the protein from its natural lipid membrane. While this process has been improved in recent years, DeLisa says, it creates a bottleneck because each protein may require a different detergent, and the technique can ruin the protein’s biological functionality.
For their SIMPLEx method, the engineers used recombinant DNA techniques to stitch together an artificial membrane protein with an identity crisis – one that maintains its biological function, but thinks it’s soluble in water.
Using E. coli as their host cells, they modified one end of their desired membrane protein with a decoy protein that tricks the E. coli into expressing the membrane protein in the cytoplasm instead of in the lipid membrane. At the other end, they attached what’s called an amphipathic protein, which has both a hydrophilic and hydrophobic end, as a shield that protects the membrane protein from water – an effective substitute for the normal role of the lipid membrane.
The technique so far appears to be applicable to a wide range of membrane proteins from all domains of life. The researchers hope to continue experimenting with it not only to fully characterize more membrane protein structures, but also to engineer proteins used in biological pathways like glycosylation, which is the ubiquitous life process of attaching sugar molecules to proteins. This is one of DeLisa’s main research interests.
The paper, titled “Making water-soluble integral membrane proteins in vivo using an amphipathic protein fusion strategy,” included co-author Lois Pollack, professor of applied and engineering physics, whose lab provided small-angle X-ray scattering characterization of the chimeric protein structures, performed at the Cornell High Energy Synchrotron Source. The work was supported by the National Science Foundation and the National Institutes of Health.
Get Cornell news delivered right to your inbox.
Subscribe