Study tracks how milk nutrients shape infant microbiome
By Krishna Ramanujan
A new study in mice helps explain why gut microbiomes of breastfed infants can differ greatly from those of formula-fed infants.
The study, “Dietary Sphinganine Is Selectively Assimilated by Members of the Mammalian Gut Microbiome,” was published in July in the Journal of Lipid Research.
The paper describes an innovative technique developed at Cornell to track the fate of metabolites – nutrients formed in or necessary for metabolism – through a mouse’s digestive tract and identify how they interact with specific gut microbes.
“We think the methods are expandable to many different microbiome systems,” said senior author Elizabeth Johnson, assistant professor of nutritional sciences in the College of Agriculture and Life Sciences. She noted that researchers investigating effects of a high-fat vs. low-fat diet, or a keto diet, might use the technique to track metabolites.
The methodology could reveal how specific metabolites promote specific bacteria. This could allow nutritionists to prescribe that patients eat foods containing specific metabolites to intentionally change the composition of their microbiomes, Johnson said.
Human milk and many other foods contain a class of lipid metabolites called sphingolipids. Previous research suggested that these metabolites help shape an infant’s microbiome, but it was not known if they actually interact with the microbiome.
The study identified two types of gut microbes, Bacteroides and Bifidobacterium, that use sphingolipids for their own metabolism.
While very little is known about the specific roles of gut microbes in human health, Bacteroides have been implicated in both beneficial and not-so-beneficial effects, depending on context. They are generally associated with microbiomes of healthy breastfed infants. Bifidobacterium, shown for the first time in this study to process dietary sphingolipids, are considered the quintessential beneficial bacteria, comprising up to 95% of breastfed infants microbiome.
They’re also a highly popular over-the-counter probiotic.
“Our lab is very interested in how the diet interacts with the microbiome in order to really understand how you can best modulate it to have positive effects on health,” Johnson said. “In this study, we were able to see that yes, these dietary lipids that are a big part of [breastfed] infants diets, are interacting quite robustly with the gut microbiome.”
Sphingolipids originate from three main sources: diet; bacteria that can produce them; and most host tissues.
Johnson, along with first author Min-Ting Lee, a doctoral student, and Henry Le, a postdoctoral researcher, both in Johnson’s lab, created a technique to specifically track dietary sphingolipids as they passed through the mouse gut.
“We custom synthesized the sphingolipid we added to the diet,” Johnson said. “It is almost identical to ones derived from breast milk but with a small chemical tag so we could trace the location of the sphingolipid once it was ingested by the mice.”
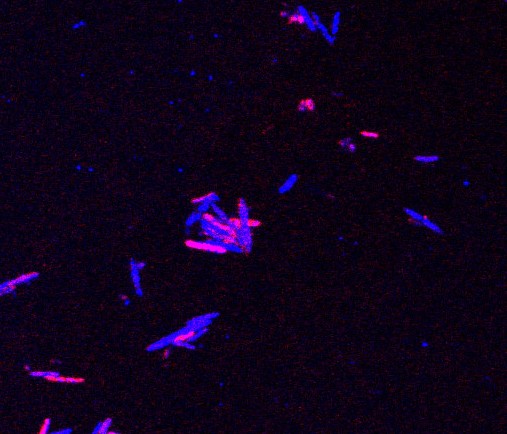
Lee then used a fluorescent label that attached to cells or microbes that absorbed the tagged lipid, such that any bacteria that had taken up sphingolipids lit up red. Microbes from the mice’s microbiomes were then isolated and analyzed. Populations with red microbes were separated from the others, and these were then genetically sequenced to identify the species of bacteria.
With further investigation, Le was able to identify the metabolites that Bacteroides and Bifidobacterium produce when exposed to dietary sphingolipids. Further investigations are underway to determine whether these microbially-produced metabolites are beneficial for infant health.
Johnson recently received a five-year, $1.9 million Maximizing Investigators’ Research Award from the National Institutes of Health (NIH) to expand on this work, to better understand how lipid-dependent host-microbe interactions affect human health..
The study was supported by seed funds from the Genomics Facility of the Biotechnology Resource Center at Cornell’s Institute of Biotechnology.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe