Magnetic tweezers reveal ‘hairballs’ in polymer growth
By Tom Fleischman
Conventional wisdom has said that when molecules known as monomers band together to create a polymer chain, that creation takes place steadily as the chain forms, like spaghetti out of a pasta maker. But a Cornell research collaboration shows that’s just not the case.
In what one reviewer called a landmark study, a group led by Peng Chen, the Peter J.W. Debye Professor of Chemistry and Chemical Biology, has achieved several firsts while offering up a new theory on polymer growth.
Using magnetic tweezers, a measurement technique never before used to study living polymerization, Chen and his group visualized growth at the single-polymer level, which also hadn’t been done before. Furthermore, the group’s study established that polymerization catalysis can be visualized at the single-molecule level under continuous turnover conditions.
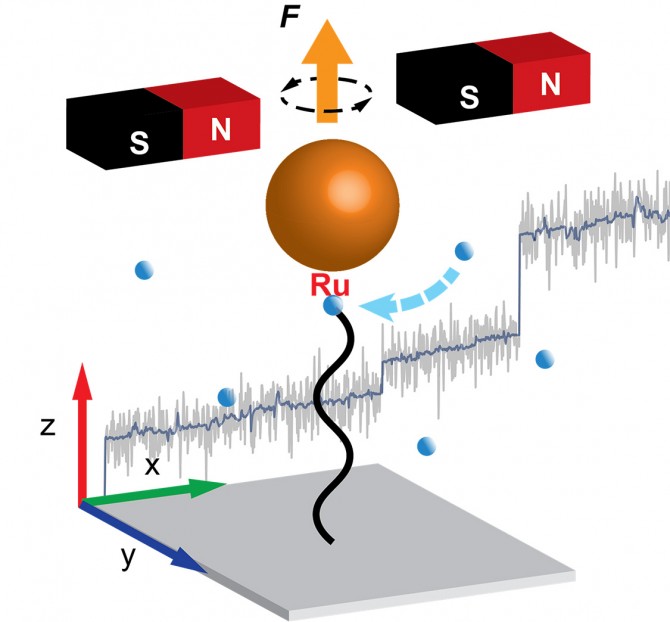
This groundbreaking work revealed the continual formation and unraveling of conformational entanglements, nicknamed “hairballs,” that produce a step-wise extension pattern of polymer growth, as opposed to more continuous extension as was previously thought.
Their findings are reported in “Single Polymer Growth Dynamics,” published Oct. 20 in Science. In addition to Chen, co-senior authors are Geoffrey Coates, the Tisch University Professor and associate chair of the Department of Chemistry and Chemical Biology, and Fernando Escobedo, the Marjorie L. Hart Professor of Engineering in the Smith School of Chemical and Biomolecular Engineering.
Lead authors are postdoctoral researcher Chunming Liu and doctoral student Kaori Kubo, both members of the Chen Group, as well as master’s student Endian Wang of the Escobedo Group.
Not only does the discovery of these “hairballs” appear to play a key role in determining the polymerization rates of individual polymers, this work could also help inform study of living bio-polymerization in cells.
In a typical polymerization reaction, a polymer chain grows from a catalyst continually to reach thousands of sub-units, but growth at the single-polymer level was not understood and hadn’t previously been observed.
“It was motivated by the long-existing knowledge that every polymer is different,” Chen said. “But people have never looked into it, and what I’ve been interested in is single-molecule studies.”
Using a so-called Grubbs catalyst, a robust complex known for its tolerance of manipulation in chemical experimentation, the group used magnetic tweezers measurements to monitor the growth of a polymer chain in real time. The measurement is done by anchoring one end of the growing polymer to a coverslip and the other to the catalyst, which is fitted with a tiny magnetic particle.
Constant application of a magnetic force pulls the particle and stretches the polymer. During polymerization, the insertion of new monomers leads to a lengthening of the polymer chain, and by tracking the position of the magnetic particle, the researchers tracked the growth of the polymer with nanometer-scale precision.
What they discovered is that, instead of steady extension growth, the polymer exhibits step-wise extension growth – the monomers do not add to the polymer extension during the growth period but instead appear to form conformational entanglements, or “hairballs.”
The periods between extension spurts are hundreds of seconds, and the spurts are up to thousands of nanometers, equivalent to thousands of monomers. Calculations showed that the polymer extension could not have grown in a steady fashion but instead in wait-and-jump steps. Simulations provided further insight into the forces that produce the hairballs and hold them together temporarily before they unravel suddenly.
“Most chemists would expect polymers to grow evenly from the active site, kind of like a string of spaghetti exiting a pasta machine,” Coates said. “What is unexpected, and therefore fascinating, is that this work shows the polymer lengthens erratically, which is attributed to the polymer chain being tangled as it leaves the active site, followed by bursts of disentanglement.”
“In fact,” Escobedo said, “the model suggests that after each monomer is inserted, the chain gets twisted a little bit, as when turning the key of a wind-up toy. After many such ‘turns’ have taken place, a portion of the newly grown polymer would become entangled like a twisted telephone cord that then takes a while to untangle.”
New experiments, Chen said, will attempt to perturb the polymerization by either introducing new interactions or disrupting them – “to either make the hairball more stable or less stable,” he said.
The group believes that bio-polymerization – such as peptide synthesis on ribosomes and polysaccharide synthesis in the cell wall – could also be explored and better understood using this technique. “It is known in biology that when the peptide comes out, it spontaneously collapses into its own compact conformation, so this is very analogous,” Chen said.
This research is another example of the type of collaboration that sets Cornell apart from many institutions.
“This is a truly collaborative project between three scientists with vastly different skill sets – a theorist [Escobedo], a physical chemist [Chen] and a synthetic chemist [Coates],” Coates said. “It seems almost impossible for this work to have been done with a subset of the expertise present.”
Part of the research was done at the Cornell Chemistry Nuclear Magnetic Resonance Core Facility, the Extreme Science and Engineering Discovery Environment, and the Cornell Center for Materials Research, which are supported by the National Science Foundation (NSF).
This work was supported by grants from the Army Research Office, the Department of Energy and the NSF.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe