William Thomas, a graduate student in the field of chemistry and chemical biology, collects data on ribonucleotide reductase.
Study offers new target for antibiotic resistant bacteria
By Linda B. Glaser
As antibiotic resistance rises, the search for new antibiotic strategies has become imperative. In 2013, the Centers for Disease Control estimated that antibiotic resistant bacteria cause at least 2 million infections and 23,000 deaths a year in the U.S.; a recent report raised the likely mortality rate to 162,044.
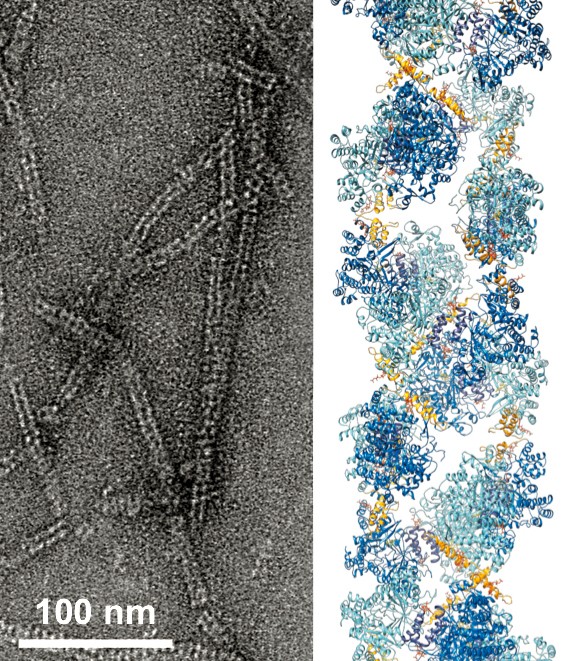
New Cornell research on an enzyme in bacteria essential to making DNA offers a new pathway for targeting pathogens. In “Convergent Allostery in Ribonucleotide Reductase,” published June 14 in Nature Communications, researchers used the MacCHESS research stations at the Cornell High Energy Synchrotron Source (CHESS) to reveal an unexpected mechanism of activation and inactivation in the protein ribonucleotide reductase (RNR).
Understanding the “switch” that turns RNR off provides a possible means to shut off the reproduction of harmful bacteria.
RNRs take ribonucleotides, the building blocks of RNA, and convert them to deoxyribonucleotides, the building blocks of DNA. In all organisms, the regulation of RNRs involves complex mechanisms, and for good reason: These mechanisms prevent errors and dangerous mutations.
“Without the RNR enzyme, DNA-based life as we know it could not exist. The particular class of RNRs that we studied in this paper is unique to bacteria, including many pathogens, which means that it’s an intriguing target for possible antibiotics,” said first author William Thomas, a graduate student in the field of chemistry and chemical biology.
“If we understand the RNR ‘off-switch’ well enough, we can take advantage of it by developing our own ways to toggle it with new antibiotic drug molecules,” Thomas said. “Notably, since this bacterial class of RNRs uses a different regulation mechanism from human RNR, we can use our newfound understanding to design drugs that target the pathogenic version without accidentally hitting the human enzyme as collateral damage.”
The research reveals evolution in action, said Nozomi Ando, assistant professor of chemistry in the College of Arts and Sciences and the paper’s senior author. The lack of the normal regulatory “switch” mechanism may actually provide an evolutionary advantage for the bacteria they studied.
“Usually the increased chance of mutations is a problem for bacteria, but maybe under certain circumstances it’s actually advantageous for an organism to mutate and possibly become resistant to an antibiotic or another stressful situation,” she said.
RNRs are not easy proteins to work with or understand, and the researchers said that characterizing them in the traditional way has been challenging.
“The combination of biological small-angle X-ray scattering using CHESS, crystallography, and cryo-electron microscopy is what made this study possible,” Ando said. “The amount of X-ray scattering data in our paper is equivalent to 11 synchrotron trips, which is unprecedented. I don’t know any structural biology paper that required that much X-ray data. But it has given us a greater understanding of the protein that had been previously impossible; we were able to map all the different shapes that it can take on, whether inactive or active, and how it switches between the two.”
Co-authors include Cornell doctoral student Audrey Burnim; research associate John-Paul Bacik; F. Phil Brooks III of Princeton University; JoAnne Stubbe of the Massachusetts Institute of Technology; Jason T. Kaelber of Rutgers University; and James Z. Chen of Oregon Health and Science University.
The research was supported by the National Science Foundation and the National Institute of Health.
Linda B. Glaser is a writer for the College of Arts and Sciences.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe