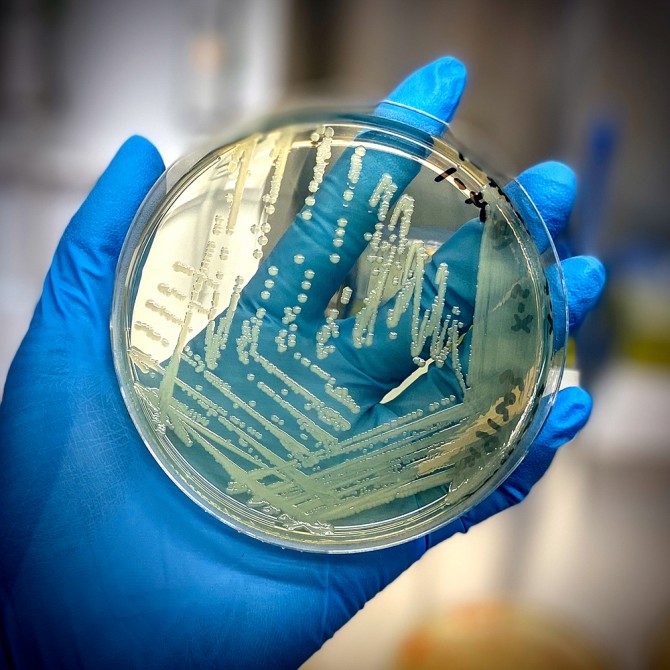
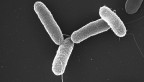
Food scientists slice time off salmonella identification process
By Blaine Friedlander
The conventional scientific process for identifying bacteria’s family – known as serotyping – can be time consuming. For salmonella, it used to take three days, and in some cases more than 12 days to assign a final classification for complex servovars.
Researchers from Cornell, the Mars Global Food Safety Center in Beijing, and the University of Georgia have developed a method for completing whole-genome sequencing to determine salmonella serotypes in just two hours and the whole identification process within eight hours.
Their research was published Feb. 24 online in the Journal Food Microbiology.
Determining salmonella’s serotype makes it easier for food safety sleuths to find the source of bacterial contamination, which can occur in a wide range of foods, such as fruits, vegetables, nuts, meat, cereal, infant formula and pet food.
“As the food supply chain becomes ever more global and interconnected, the opportunity for food to become contaminated with salmonella increases,” said lead author Silin Tang, Ph.D. ’15, senior research scientist in microbial risk management at the Mars Global Food Safety Center in China. “In the fast-moving world of food manufacturing, where rapid identification and response to salmonella contamination incidents is critical, developing a more efficient pathogen identification method is essential.”
Conventional serotyping has been at the core of public health monitoring of salmonella infections for a half-century, Tang said. But long turnaround times, high costs and complex sample preparations have led global food safety regulators, food authorities and public health agencies to change to whole-genome sequencing methods for pathogen subtyping.
All 38 salmonella strains – representing 34 serotypes – assessed in this study were accurately predicted to the serotype level using whole-genome sequencing.
This is important news for the food industry, as very few laboratories can conduct classical serotyping, said Martin Wiedmann, Ph.D. ’97, the Gellert Family Professor in Food Safety and a Cornell Institute for Food Systems faculty fellow.
“In some countries,” Wiedmann said, “it can take up to two days to even get the suspected salmonella to a certified lab.”
With whole genome sequencing, he said, the new state-of-the-art test relies on simple equipment. “For the food industry, processing plants are in the middle of nowhere,” he said. “Now you can conduct testing in a lab that’s close to the food processing plant.”
In the United States, salmonella bacteria cause approximately 1.3 million infections, more than 26,000 hospitalizations and more than 400 deaths annually, according to annual estimates from the U.S. Centers for Disease Control and Prevention.
Different salmonella serotypes often come from different places, said Wiedmann, who explained the process through an example: chicken pot pie.
“Where does the salmonella come from?” he said. “You have carrots, peas, obviously chicken, and spices. If you have salmonella enteritidis – that’s usually associated with chicken – then you look for and track down the source of the chicken. If you have salmonella virchow, the serotype usually associated with food in southeast Asia, then you want to track down the spices from there. … Serotyping provides food safety scientists with a priority list of where to look.”
In addition to Silin and Wiedmann, the other researchers on the study, “Evaluation of Real-Time Nanopore Sequencing for Salmonella Serotype Prediction,” are Feng Xu, Chongtao Ge, Hao Luo, Guangtao Zhang, Abigail Stevenson and Robert C. Baker of the Mars Global Food Safety Center; and Shaoting Li and Xiangyu Deng of the University of Georgia. Funding was provided by the Mars Global Food Safety Center.
Media Contact
Get Cornell news delivered right to your inbox.
Subscribe